Suffix Table
\( \def\sa#1{\tt{#1}} \def\tsuff{\mathit{suff}} \def\dd{\dot{}\dot{}} \)Let $x$ be a non-empty string. The suffix table of $x$ is defined on its positions $i$, $i=0, \dots, |x|-1$, by: $\tsuff[i]$ is the length of the longest suffix of $x$ ending at position $i$. Obviously $\tsuff[[x|-1]=|x|$.
Here is the suffix table of the word $\sa{abaababaaba}$:
| $i$ | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| $x[i]$ | $\sa a$ | $\sa b$ | $\sa a$ | $\sa a$ | $\sa b$ | $\sa a$ | $\sa b$ | $\sa a$ | $\sa a$ | $\sa b$ | $\sa a$ |
| $\tsuff[i]$ | 1 | 0 | 3 | 1 | 0 | 6 | 0 | 3 | 1 | 0 | 11 |
It can be computed in $O(|x|)$ time by the following algorithm.
\begin{algorithmic}
\STATE $suff[m-1]\leftarrow 1$
\STATE $g\leftarrow m-1$
\FOR{$i\leftarrow m-2$ \DOWNTO $0$}
\IF{$i\gt g \mbox{ and } suff[i+m-1-f]\ne i-g$}
\STATE $suff[i]\leftarrow \min\{suff[i+m-1-f],i-g\}$
\ELSE
\STATE $(f,g)\leftarrow (i,\max\{i,g\})$
\WHILE{$g\ge 0$ and $x[g]=x[g+m-1-f]$}
\STATE $g\leftarrow g-1$
\ENDWHILE
\STATE $suff[i]\leftarrow f-g$
\ENDIF
\ENDFOR
\RETURN{$suff$}
\end{algorithmic}
The key idea in Algorithm Suffixes that computes the table sequentially from right to left is to benefit from what has been computed before the current position.
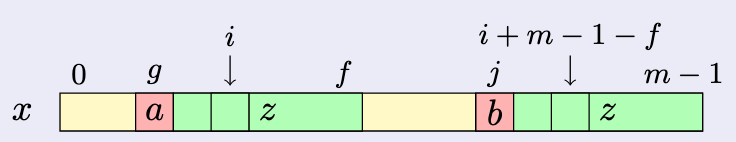
When $z=x[g+1\dd f]$ is a suffix of $x$ and position $i$ is between $g$ and $f$ (see picture), the first step for computing $\tsuff[i]$ is to check whether or not its value can be deduced from the work done on the suffix occurrence of $z$, that is, at position $i+m-1-f$ on $x$. This saves enough work to get a linear-time algorithm.